GCP – Building a genomics analysis architecture with Hail, BigQuery, and Dataproc
We hear from our users in the scientific community that having the right technology foundation is essential. The ability to very quickly create entire clusters of genomics processing, where billing can be stopped once you have the results you need, is a powerful tool. It empowers the scientific community to spend more time doing their research and less time fighting for on-prem cluster time and configuring software.
At Google Cloud, we’ve developed healthcare-specific tooling that makes it easy for researchers to look at healthcare and genomic data holistically. Combining genotype data with phenotype data from electronic health records (EHRs), device data, medical notes, and medical images makes scientific hypotheses limitless. And, our analysis platforms like AI Platform Notebooks and Dataproc Hub let researchers easily work together using state-of-the-art ML tools and combine datasets in a safe and compliant manner.
Building an analytics architecture for genomic association studies
Genome-wide association studies (GWAS) are one of the most prevalent ways to study which genetic variants are associated with a human trait, otherwise known as a phenotype. Understanding the relationships between our genetic differences and phenotypes such as diseases and immunity is key to unlocking medical understanding and treatment options.
Historically, GWAS studies were limited to phenotypes gathered during a research study. These studies were typically siloed, separate from day-to-day clinical data. However, the increased use of EHRs for data collection, coupled with natural language processing (NLP) advances that unlock the data in medical notes, has created an explosion of phenotype data available for research. In fact, Phenome-wide association studies (PheWas) are gaining traction as a complementary way to study the same associations that GWAS provides, but starting from the EHR data.
In addition, the amount of genomics data now being created is causing storage bottlenecks. This is especially relevant as clinical trials move toward the idea of basket trials, where patients are sequenced for hundreds of genes up front, then matched to a clinical trial for a gene variant.
While all of this data is a boon for researchers, most organizations are struggling to provide their scientists with a unified platform for analyzing this data in a way that balances respecting patient privacy with sharing data appropriately with other collaborators.
Google Cloud’s data lake empowers researchers to securely and cost-effectively ingest, store, and analyze large volumes of data across both genotypes and phenotypes. When this data lake infrastructure is combined with healthcare-specific tooling, it’s easy to store and translate a variety of healthcare formats, as well as reduce toil and complexity. Researchers can move at the speed of science instead of the speed of legacy IT. A recent epidemiology studycited BigQuery as a “cloud-based tool to perform GWAS,” and suggests that a future direction for PheWAS “would be to extend existing [cloud platform] tools to perform large-scale PheWAS in a more efficient and less time-consuming manner.”
The architecture we’ll describe here offers one possible solution to doing just that.
GWAS/PheWAS architecture on Google Cloud
The goal of the below GWAS/PheWAS architecture is to provide a modern data analytics architecture that will:
-
Safely and cost-effectively store a variety of large-scale raw data types, which can be interpreted or feature-engineered differently by scientists depending on their research tasks
-
Offer flexibility in analysis tools and technology, so researchers can choose the right tool for the job, across both Google Cloud and open source software
-
Accelerate the number of questions asked and increase the amount of scientific research that can be done by:
-
Reducing the time scientists and researchers spend implementing and configuring IT environments for their various tools
-
Increasing access to compute resources that can be provisioned as needed
-
Make it easy to share and collaborate with outside institutions while maintaining control over data security and compliance requirements.
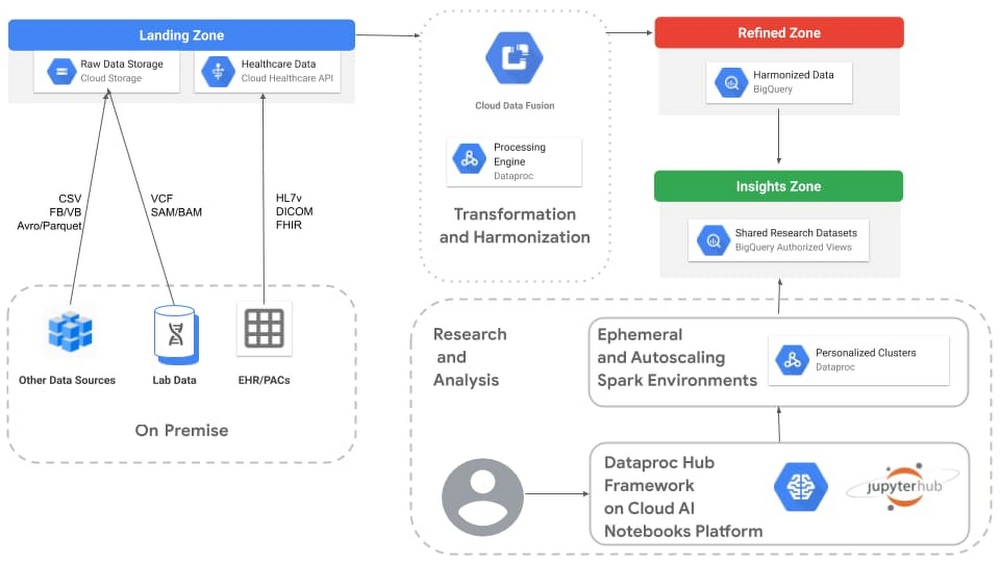
Check out full details on our healthcare analytics platform, including a reference architecture. The architecture depicted below represents one of many ways to build a data infrastructure on Google Cloud. The zones noted in the image are logical areas of the platform that make it easier to explain the purpose for each area. These logical zones are not to be confused with Google Cloud’s zones, which are physical definitions of where resources are located.
This particular architecture is designed to enable data scientists to perform GWAS and PheWAS analysis using Hail, Dataproc Hub, and BigQuery.
Here’s more detail on each of the components.
Landing zone
The landing zone, also referred to by some customers as their “raw zone,” is where data is ingested in its native format without transformations or making any assumptions about what questions might be asked of it later.
For the most part, Cloud Storage is well-suited to serve as the central repository for the landing zone. It is easy to bring genomic data stored in raw variant call format (VCF) or SAM/BAM/CRAMfiles into this durable and cost-effective storage. A variety of other sources, such as medical device data, cost analysis, medical billing, registry databases, finance, and clinical application logs are also well suited for this zone, with the potential to be turned into phenotypes later. Take advantage of storage classes to get low-cost, highly durable storage on infrequently accessed data.
For clinical applications that use the standard healthcare formats of HL7v2, DICOM, and FHIR, the Cloud Healthcare API makes it easy to ingest the data in its native format and tap into additional functionality, such as:
-
Automated de-identification
-
Direct exposure to the AI Platform for machine learning
-
Easy export into BigQuery, our serverless cloud data warehouse
Transformation and harmonization
The goal of this particular architecture is to prepare our data for use in BigQuery. Cloud Data Fusion has a wide range of prebuilt plugins for parsing, formatting, compressing, and converting data. Cloud Data Fusion also includes Wrangler, a visualization tool that interactively filters, cleans, formats, and projects the data, based on a small sample (1000 rows) of the dataset. Cloud Data Fusion generates pipelines that run on Dataproc, making it easy to extend Data Fusion pipelines with additional capabilities from the Apache Spark ecosystem. Fusion can also help track lineage between the landing and refined zones.
For a more complete discussion of preparing health data for BigQuery, check out Transforming and harmonizing healthcare data for BigQuery.
Direct export to BigQuery
BigQuery is used as the centerpiece of our refined and insights zones, so many healthcare and life science formats can be directly exported into BigQuery. For example, a FHIR store can be converted to a BigQuery dataset with a single command line call of
gcloud beta healthcare fhir-stores export bq.
See this tutorial for more information on ingesting FHIR to BigQuery.
When it comes to VCF files, the Variant Transforms tool can load VCF files from Cloud Storage into BigQuery. Under the hood, this tool uses Dataflow, a processing engine that can scale to loading and transforming hundreds of thousands of samples and billions of records. Later in this post, we’ll discuss using this Variant Transforms tool to convert data back from BigQuery and into VCF.
Refined zone
The refined zone in this genomics analysis architecture contains our structured, yet somewhat disconnected data. Datasets tend to be associated with specific subject areas but standardized by Cloud Data Fusion to use specific structures (for example, aligned on SNOWMED, single VCF format, unified patient identity, etc). The idea is to make this zone the source of truth for your tertiary analysis.
Since the data is structured, BigQuery can store this data in the refined zone, but also start to expose analysis capabilities, so that:
-
Subject matter experts can be given controlled access to the datasets in their area of expertise
-
ETL/ELT writers can use standard SQL to join and further normalize tables that combine various subject areas
-
Data scientists can run ML and advanced data processing on these refined datasets using Apache Spark on Dataproc via the BigQuery connector with Spark.
Insights zone
The insights zone is optimized for analytics and will include the datasets, tables, and views designed for specific GWAS/PheWAS studies.
BigQuery authorized views lets you share information with specified users and groups without giving them access to the underlying tables (which may be stored in the refined zone). Authorized views is often an ideal way to share data in the insights zone with external collaborators.
Keep in mind that BigQuery (in both the insights and refined zones) offers a separation of storage from compute, so you only need to pay for the processing needed for your study. However, BigQuery still provides many of the data warehouse capabilities that are often needed for a collaborative insights zone, such as managed metadata, ACID operations, snapshot isolation, mutations, and integrated security. For more on how BigQuery storage provides a data warehouse without the limitations associated with traditional data warehouse storage, check out Data warehouse storage or a data lake? Why not both?
Research and analysis
For the actual scientific research, our architecture uses managed Jupyter Lab notebook instances from AI Platform Notebooks. This enterprise notebook experience unifies the model training and deployment offered by AI Platform with the ingestion, preprocessing, and exploration capabilities of Dataproc and BigQuery.
This architecture uses Dataproc Hub, which is a notebook framework that lets data scientists select a Spark-based predefined environment that they need without having to understand all the possible configurations and required operations. Data scientists can combine this added simplicity with genomics packages like Hail to quickly create isolated sandbox environments for running genomic association studies with Apache Spark on Dataproc.
To get started with genomics analysis using Hail and Dataproc, check out part two of this post.
Read More for the details.